Research
Chaperone/Usher Pathway Pili
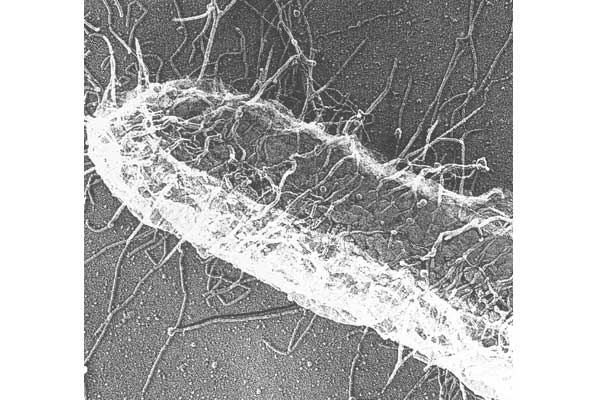
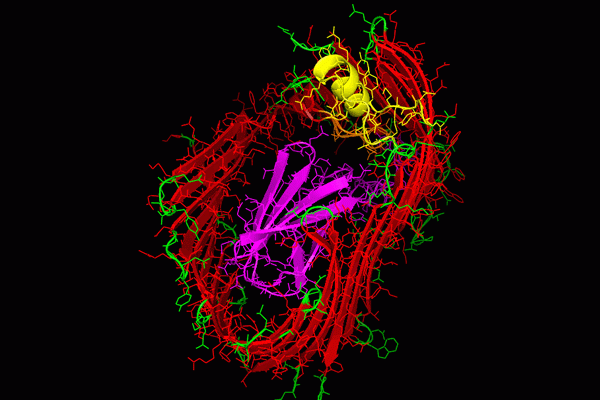
CUPs are ubiquitous in Gram-negative pathogens and are known to be critical virulence factors in a wide range of pathogenic bacterial genera, including Escherichia, Klebsiella, Acinetobacter, Pseudomonas, Haemophilus, Salmonella and Yersiniae. We have used the type 1 pili and P pili systems, which enable UPEC to cause bladder and kidney infections, respectively, as models to understand the assembly of CUP pili. We first discovered that P pili (and later type 1 and S pili) were multicomponent fibers, each consisting of a helical rod joined end to end to a linear tip fibrillum, which contains a two-domain adhesin at its distal end. Elucidating how these structures are built is uncovering basic principles of protein folding, chaperone function and macromolecular assembly.
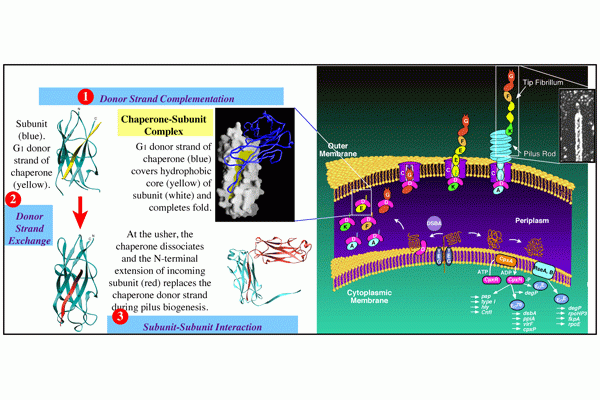
CUP Pilus biogenesis: We have used both type 1 and P pili CUP systems as models to elucidate molecular details of how the cognate chaperones and ushers, which are encoded in operons along with pilin subunits and tip adhesins, orchestrate the folding of each pilin subunit and the ordered assembly of CUP pili at the outer membrane (OM) without the input of cellular energy.
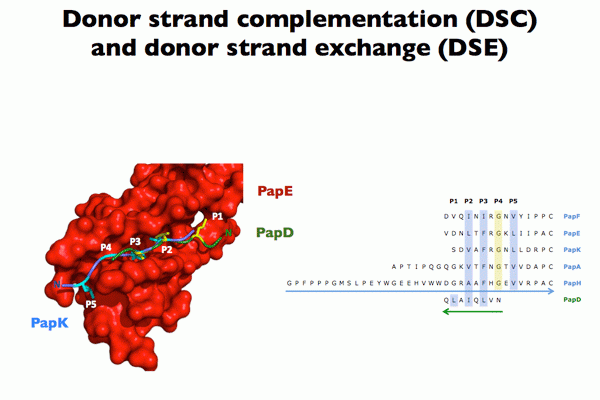
Chaperone function: We discovered that the molecular basis of chaperone-assisted folding occurred by a reaction we termed donor strand complementation (DSC). The crystal structure of the P pilus PapD chaperone revealed that it consisted of two immunoglobulin (Ig) domains. Each pilin subunit is an incomplete Ig-like domain that is missing its C-terminal beta-strand. We found that the chaperone templates folding of each of the pili subunits by donating its G1 strand such that chaperone’s alternating exposed hydrophobic residues on the G1 strand are buried in complementary pockets in the subunit completing a non-canonical Ig-like subunit fold (consisting of 6 b-strands from the subunit and the G1 chaperone b-strand serving as the 7th b-strand). The crystal structures of each of the five single-domain Ig-like P pilins and the Saf pilins in complexes with their respective chaperones revealed that subunits are held by the chaperone in a primed high-energy state having incompletely collapsed hydrophobic cores. Our studies showed that these primed complexes are differentially targeted to the OM assembly platform, we named the usher, which functions as a gated channel that catalyzes pilus assembly by driving subunit polymerization in a reaction we termed donor strand exchange (DSE). All subunits, excluding tip adhesins, contain a short N-terminal extension (Nte) comprised of conserved alternating hydrophobic residues (analogous, but not identical, to those on the chaperone G1 strand). DSE occurs in a zip-in-zip-out mechanism when an incoming subunit’s Nte replaces the G1 b-strand of the chaperone to complete the Ig fold (in a canonical fashion) of a nascently incorporated subunit at the growing terminus of the pilus resulting in chaperone dissociation and consolidation of the hydrophobic core. Thus, every subunit in the pilus completes the Ig fold of its neighbor. Energy for pilus assembly is derived in part by this stepwise folding process.
Usher function: The OM usher, that catalyzes DSE, is comprised of five functional domains: a 24-stranded integral β-barrel translocation domain (TD), a β-sandwich Plug domain (PD) that gates the pore of the TD, a periplasmic amino-terminal domain (NTD), and two carboxy-terminal domains (CTD1 and CTD2). Coordination of these domains ensures that subunits interact productively during fiber polymerization. The existing apo-structures of the type 1 and P pilus usher proteins, FimD and PapC, respectively, revealed that the PD resides within the TD lumen and blocks the pore in the absence of incoming chaperone-subunit complexes [inactive state], thus gating the TD pore and preventing large molecules from flowing freely across the OM. Pilus assembly typically begins with recruitment of the chaperone-adhesin complex to the NTD of the usher. The CTD then interacts with the NTD-chaperone-adhesin complex and the chaperone-adhesin complex transfers to the CTD and the NTD becomes free for the next chaperone-subunit complex to bind. Sometime during this process the PD translocates out of the TD pore and forms a complex with the NTD. Once the PD leaves the TD, the TD is free to go from a kidney shaped conformation to a circular form that likely facilitates the extrusion of folded pilins across the OM. The usher likely catalyzes DSE by positioning the chaperone-adhesin complex bound to the CTD and the next chaperone-subunit complex bound to the NTD in an optimal position for the interaction of the incoming subunits NTE to displace the chaperone from the adhesin and complete the final folding of the adhesin’s pilin domain. Multiple rounds of chaperone-subunit binding and DSE reactions lead to the final pilus structure, which is extruded through the usher TD pore to the extracellular surface of the bacteria.
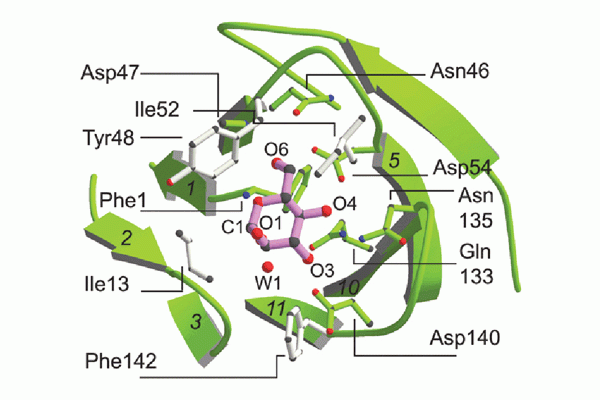
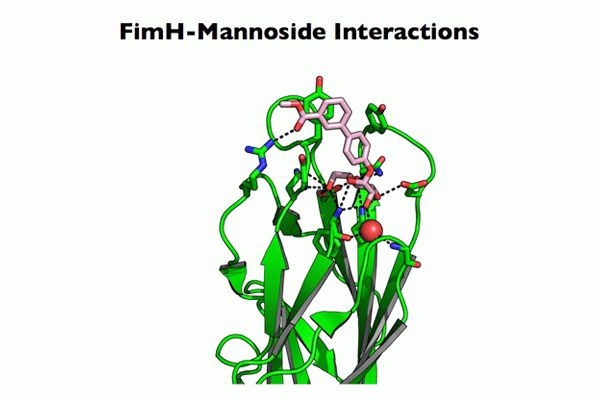
CUP adhesins: Distal CUP pilus adhesins are two-domain proteins, consisting of an N terminal receptor-binding domain and a C terminal pilin domain. The N terminal receptor-binding domain binds to host or environmental moieties, and thus has important influences on bacterial tropism and initiation of infections. The pilin domain links the receptor-binding domain to the tip of the pilus via donor strand exchange. We have solved and published the structures of multiple CUP pilus adhesins including the FimH, type 1 pilus adhesin; the PapG, P pilus adhesin; the FmlH, F9 pilus adhesin; and the UclD, F17-like adhesin. UPEC use type 1 pili tipped with the FimH adhesin, which binds mannosylated glycoproteins on the lumenal surface of the bladder to facilitate colonization. In brief, FimH-mannose interactions have been shown to be of critical importance in the ability of UPEC and other Enterobacteriaceae, such as Klebsiella, to cause UTIs. UPEC also use the P pilus tipped with the galabiose binding adhesin PapG to mediate pyelonephritis by binding to receptors in the kidney. Further, UPEC can form a reservoir in gastrointestinal tract (GIT) from which they are shed in the feces and subsequently colonize the periurethral area. We have shown that FimH/type 1 pili and F17-like pili tipped with the UclD adhesin are important in UPEC GIT colonization. Further, we have identified a role for F9 pili tipped with the galactose binding FmlH adhesin, in binding to the inflamed bladder epithelium during chronic cystitis. In addition, we have demonstrated that Acinetobacter baumannii, an emerging multidrug resistant pathogen, uses CUP pili tipped with the fibrinogen binding CupD adhesin to mediate catheter associated UTIs.
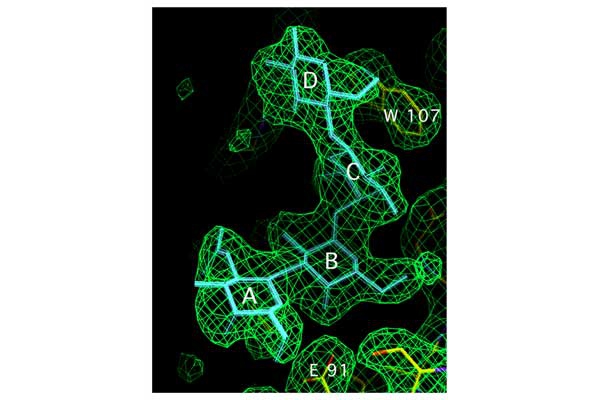
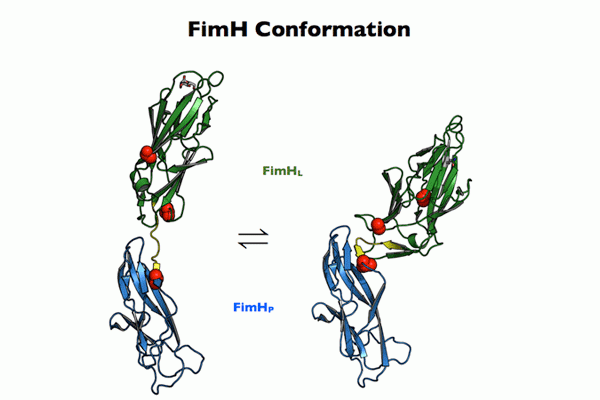
In addition to our elucidation of structures, functions and ligands of CUP adhesins, we have also assessed the evolution and conformational equilibrium of CUP adhesins. Through the analysis of multiple FimH sequences from E. coli isolates combined with functional binding and bladder infections studies, we discovered that pathoadaptive alleles of FimH with variant residues outside the mannose binding pocket, affect transitions between low and high-affinity FimH conformations and impact FimH-mediated pathogenesis. In vitro binding studies revealed that while all pathoadaptive variants tested displayed the same high affinity for mannose when bound by the chaperone FimC, affinities varied when FimH was incorporated into pilus tip structures. Structural studies have shown that FimH adopts an elongated conformation when complexed with FimC, but when incorporated into the pilus tip, FimH can also adopt a compact conformation. Interestingly, FimH variants, which maintain a high-affinity conformation in the pilus tip were attenuated during chronic bladder infection arguing that FimH’s ability to switch between conformations is important in pathogenesis. Further work to understand possible conformer equilibriums within CUP adhesins, as well as the specificity of the myriad of CUP adhesins encoded by bacteria will give important insights into bacterial tropism and disease initiation.
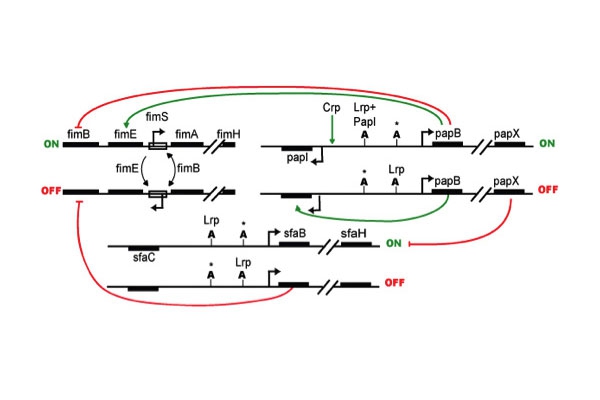
CUP regulation/cross-talk: Studies on type 1, S and P pili have demonstrated multiple regulatory elements controlling their expression. Global transcriptional regulators are known to affect multiple CUP operons. Further, each CUP operon appears to have specific associated regulators, which impact expression. Both type 1 and P pili are regulated by a process termed phase variation, where individual bacteria have all-or-none expression of pili despite the population being subject to the same environmental conditions. The fim operon is under phase variable expression regulated by site specific recombinases (FimB, FimE, FimX and others), which can switch the orientation of the fim promoter (fimS) by acting on 9bp palindromic sequences flanking fimS. In contrast, pap phase variable switching is governed by differential DAM-DNA methylation at two GATC sites, which alters regulator binding. Additional studies confirm these general themes for the regulation and function of other CUP operons as well as provide evidence that under any given condition each bacterium likely only expresses one CUP system at a time, perhaps to insure fidelity of assembly and outer membrane integrity. This apparent hierarchy of CUP expression argues for cross-regulation between the CUP operons. We are characterizing the expression of all CUP operons in a prototypical UPEC isolate, UTI89, under a variety of growth conditions and in a variety of mutants in general regulators. This work will provide insights into bacterial mechanisms that determine tropism and potential compensatory mechanisms that UPEC may use against chemotherapeutics that target a particular CUP system.
Collaborators: Han Remaut, Carl Frieden, Peng Yuan